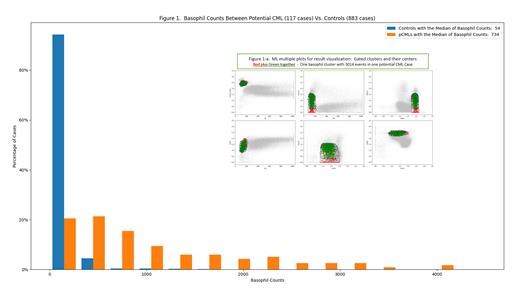
Background: In marrow, basophil counts are low but may increase in reactive or neoplastic conditions including chronic myeloid leukemia (CML). The current basophil enumeration by the gating approach is operator dependent with variable accuracy. Objective: To enumerate basophils and to compare basophil and granulocyte counts between potential CML cases (pCMLs) and controls using machine learning (ML) models. Research Methods: pCMLs and controls (117 and 883 marrow samples, respectively) were selected using key-word search from flow-cytometry (FC) reports. In general, pCMLs had both granulocytosis and basophilia pending tests for t(9;22). The datasets were from an antibody panel - FS_SS_HLA-DR_CD33_CD34_CD117_CD45. Three preliminary models, the basophil enumeration model (BEM), granulocyte enumeration model (GEM), and granulocyte-basophil model (GBM) were used (data not published). GBM incorporates basophil and granulocyte counts from BEM and GEM, and separates the cases into two groups, GB-1 with high granulocyte and basophil counts and GB-2 with low basophil and/or granulocyte counts. Results: BEM revealed higher basophil counts in pCMLs than those in the controls (median: 734 vs. 54). However, 35 (5%) of the controls had >390 basophils (≥25 percentile in pCMLs) (Figure-1). GBM classified 96 (90%) pCMLs as GB-1 and 11 as GB-2. Conversely, GBM classified 74 (10%) controls as GB-1 and the remaining 672 as GB-2 (Figure-2). Multiple scatter plots in figure 1-a illustrate two-dimensional distributions of a basophil cluster in a pCML case (green: CD117-positive basophils [often increased in pCML]; red: CD117-negative basophils). Conclusion: In experimental setting, BEM is an efficient high-throughput alternative for basophil enumeration with high consistency and is opterator-independent. GBM could separate pCMLs from most controls automatically. Future ML models will use confirmed CML-cases and additional antibody markers in a multicolor FC platform.
Disclosures
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal